Counting hierarchical kinds
« previous post | next post »
Nick Collins, "Earth is home to 8.7 million species", The Telegraph 8/23/2011:
Previous guesses had put the total number of different species at anywhere between three million and 100 million, but a new calculation based on the way in which life forms are classified puts the estimate at the lower end of that scale.
The list of known species currently stands at about 1.2 million, but experts said that advances in technology meant that the remainder could be found and classified within the next century.
The study was undertaken by researchers from the Census of Marine Life, a ten-year project involving 2,700 scientists from more than 80 countries aimed at assessing the diversity of life in our seas and oceans which concluded in October 2010.
The paper behind this story is Camilo Mora, Derek P. Tittensor, Sina Adl, Alastair G. B. Simpson, Boris Worm, "How Many Species Are There on Earth and in the Ocean?", PLoS Biology 9(8) 2011. It adds a new twist to an estimation problem whose history extends well back into the 20th century.
A classic discussion is RA Fisher, AS Corbet, and CB Williams, "The relation between the number of species and the number of individuals in a random sample of an animal population", Journal of Animal Ecology 1943. And a bit earlier, Alan Turing had made a brilliant contribution to solving a superficially different problem: Fisher et al. wanted to estimate the population distribution of butterfly species from samples gathered by lepidopterists in Malaya, while Turing and his colleagues wanted to estimate the population distribution of German letter-sequences or words from samples of plaintext messages gathered in the course of the Enigma decryption project at Bletchley Park.
Since Turing's idea was part of a classified project, it was published only in disguised form, and more than a decade later, in IJ Good, "The Population Frequencies of Species and the Estimation of Population Parameters", Biometrika 40(3-4) 1953:
A random sample is drawn from a population of animals of various species. (The theory may also be applied to studies of literary vocabulary, for example.) If a particular species is represented r times in the sample of size N, then r/N is not a good estimate of the population frequency, p, when r is small. Methods are given for estimating p, assuming virtually nothing about the underlying population. The estimates are expressed in terms of smoothed values of the numbers nr (r = 1, 2, 3, …), where nr is the number of distinct species that are each represented r times in the sample. (nr may be described as `the frequency of the frequency r'.) Turing is acknowledged for the most interesting formula in this part of the work.
If you'd like to know what that "most interesting formula" was, and how it works, see my lecture notes "Statistical estimation for Large Numbers of Rare Events"; or William Gale and Geoffrey Sampson, "Good‐Turing frequency estimation without tears", Journal of Quantitative Linguistics 1995. For a comparison of Fisher's method with the Good-Turing approach, see Bradley Efron and Ronald Thisted, "Estimating the number of unseen species: How many words did Shakespeare know?", Biometrika 1976:
Shakespeare wrote 31534 different words, of which 14376 appear only once, 4343 twice, etc. The question considered is how many words he knew but did not use. A parametric empirical Bayes model due to Fisher and a nonparametric model due to Good & Toulmin are examined. The latter theory is augmented using linear programming methods. We conclude that the models are equivalent to supposing that Shakespeare knew at least 35000 more words.
And for an extensive, systematic and clear exploration of the modeling issues in the textual case, see Harald Baayen, Word Frequency Distributions, 2001. Some applications in other areas can be found in Iuliana Ionita-Laza, Christoph Lange and Nan M. Laird, "Estimating the number of unseen variants in the human genome", PNAS 2009; "Vicki Pollard's Revenge", Language Log 1/2/2007; "Comparing the vocabularies of different languages", Language Log 3/31/2008
Given that background, let's take a look at the new twist in Camilo Mora, Derek P. Tittensor, Sina Adl, Alastair G. B. Simpson, Boris Worm, "How Many Species Are There on Earth and in the Ocean?", PLoS Biology 9(8) 2011. Here's an abridged version of their introduction:
Robert May recently noted that if aliens visited our planet, one of their first questions would be, “How many distinct life forms—species—does your planet have?” He also pointed out that we would be “embarrassed” by the uncertainty in our answer. This narrative illustrates the fundamental nature of knowing how many species there are on Earth, and our limited progress with this research topic thus far. Unfortunately, limited sampling of the world's biodiversity to date has prevented a direct quantification of the number of species on Earth, while indirect estimates remain uncertain due to the use of controversial approaches […]. Globally, our best approximation to the total number of species is based on the opinion of taxonomic experts, whose estimates range between 3 and 100 million species […]. With the exception of a few extensively studied taxa […], we are still remarkably uncertain as to how many species exist, highlighting a significant gap in our basic knowledge of life on Earth. Here we present a quantitative method to estimate the global number of species in all domains of life. We report that the number of higher taxa, which is much more completely known than the total number of species, is strongly correlated to taxonomic rank and that such a pattern allows the extrapolation of the global number of species for any kingdom of life.
So their basic idea is to generalize the problem in a hierarchical way. Earlier authors addressed the problem of estimating the distribution of individuals in species, but also noted in passing that similar issues arise in relating species to genera, or genera to families, and so on. Mora et al. look at this hierarchical generalization in a systematic way.
They add one additional twist. The methods discussed up to this point have been based on the concept of extrapolating a type-token plot like this one:

Here the horizontal axis is the number of word tokens examined in running text, while the vertical axis is the number of word types (distinct letter sequences) found. The Fisher paper framed things in terms of the number of species (animal types) found, as a function of the number of individual animals (animal tokens) examined.
Rather than considering the number of higher-level kinds found as a function of the number of lower-level entities examined, Mora et al. relate the number of discovered higher-level kinds to the simple passage of historical time (what they call a "temporal accumulation curve". If we assume that individual words (or animals) are examined at a constant rate, then this is the same thing with a change of scale — and they devote some effort to estimating the sensitivity of their estimates to plausible variation over time in the amount of such taxonomic effort that is expended.
Whether they've done enough to lay such concerns to rest is controversial. Thus Carl Zimmer ("How Many Species on Earth? It’s Tricky", NYT 8/23/2011) quotes Terry Erwin as objecting “They’re measuring human activity, not biodiversity.”
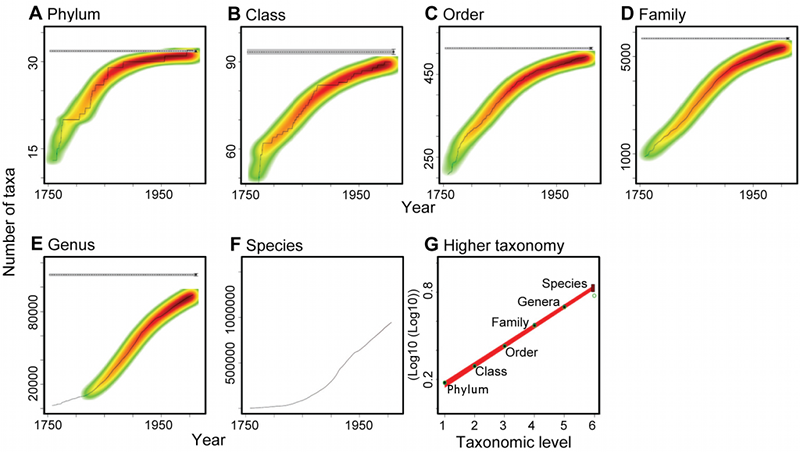
Anyhow, the higher we go in the tree of taxa, the closer the current observational estimates of the number of types are to the asymptotes that they will eventually reach as more and more of the relevant populations are sampled. Here's Mora et al.'s Figure 1, showing the growth over historical time, from the mid-18th-C to the present, of estimates of the numbers of phyla, classes, orders, etc.
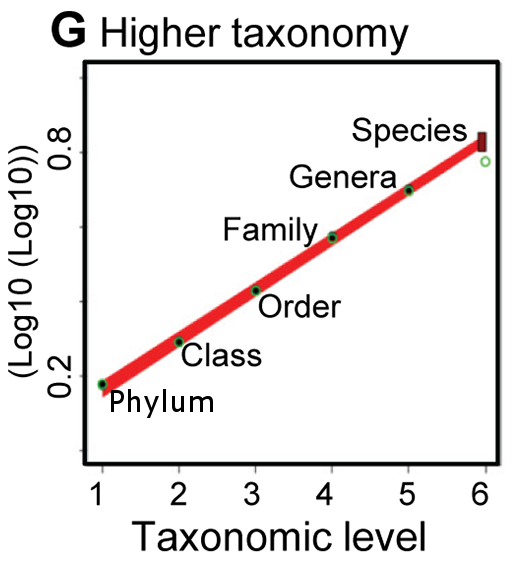
(A–F) The temporal accumulation of taxa (black lines) and the frequency of the multimodel fits to all starting years selected (graded colors). The horizontal dashed lines indicate the consensus asymptotic number of taxa, and the horizontal grey area its consensus standard error. (G) Relationship between the consensus asymptotic number of higher taxa and the numerical hierarchy of each taxonomic rank. Black circles represent the consensus asymptotes, green circles the catalogued number of taxa, and the box at the species level indicates the 95% confidence interval around the predicted number of species.
And this turns out to be crucial, because Panel F, plotting the count of species as a function of time, is clearly far away from its asymptotic value — which they don't try to estimate directly. The key part of their approach is indicated in Panel G, which plots the log of the log of the estimated asymptotes as a function of taxonomic level:
More specifically,
… we accounted for undiscovered higher taxa by fitting, for each taxonomic level from phylum to genus, asymptotic regression models to the temporal accumulation curves of higher taxa (Figure 1A–1E) and using a formal multimodel averaging framework based on Akaike's Information Criterion to predict the asymptotic number of taxa of each taxonomic level (dotted horizontal line in Figure 1A–11E). Secondly, the predicted number of taxa at each taxonomic rank down to genus was regressed against the numerical rank, and the fitted models used to predict the number of species (Figure 1G).
[Of course, error bars shrink marvelously when expressed in terms of the log of the log of the basic numbers at issue…]
One obvious question is whether there's any effective way to apply this idea to the problems of linguistic frequency estimation.
peter said,
August 24, 2011 @ 8:29 am
Is the "Toulmin" mentioned alongside Good in the quote from Efron and Thisted the philosopher of argumentation Stephen E. Toulmin, or is that a mis-typing of "Turing"?
[(myl) Neither one. The reference is I.J. Good & G.H. Toulmin, "The number of new species, and the increase in population coverage, when a sample is increased", Biometrika 1956.]
Jonathan Badger said,
August 24, 2011 @ 8:39 am
It isn't even clear if there's an effective way to apply this in its own domain of biology. As Jonathan Eisen points out in his blog, the microbial estimates are frankly absurd, which should give one pause.
[(myl) But given the lack of tree-structured descent in bacteria and archaea, due to rampant borrowing of DNA, I've never understood how the concept of "species" even applies there. Does this problem really affect the rest of their analysis?]
peter said,
August 24, 2011 @ 8:43 am
For biological applications, I wonder to what extent these estimation methods each suffer from the fact that our biological taxa are socially constructed. For instance, there are about 35 species in the genus malus (apple trees) and a similar number of species in the genus pyrus (pear trees), yet over 700 species in the genus Eucalyptus, because the initial classifications were undertaken by 18th-century Europeans and not by Aboriginal Australians. As a consequence, one genus may contain much greater numbers and diversity of species than another.
[(myl) My outsider's impression is that there is much better agreement about what the overall descent-tree is (at least among animals and plants that don't swap DNA so promiscuously that a tree makes no sense) than about how to label the nodes of the tree with taxonomic level.
If that's true, then there are two interpretations of the Mora et al. method. One depends on assuming that variation in assigning node-type labels (sub-species, species, genus, famliy, etc.) will average out so that different decisions won't change the overall shape of the relationships (though consistent splitters and lumpers would wind up with different quantitative results). The other interpretation would agree that we're modeling a social process — specifically, we're trying to predict what will happen over time if systematic biology continues to unfold in the future as it has in the past.]
Jonathan Badger said,
August 24, 2011 @ 9:21 am
Many of the same issues with microbial species also apply to other groups of organisms such as plants and fungi to a lesser degree. But more importantly, as far we can tell, the *majority* of biodiversity is microbial. To give an analogy, would you trust a linguistic diversity estimate that obviously failed for Niger–Congo and Austronesian languages just because it came up with a not implausible number for Indo-European?
[(myl) No — but the uniformitarian hypothesis seems to have a much stronger presumption of truth with respect to the development of human language families, than with respect to the biological history of bacteria as opposed to (say) insects or fish.
I'd turn your question around, as follows: Does the method of Mora et al. appear to improve our estimate of the number of insect or fish species?]
Carl Zimmer said,
August 24, 2011 @ 10:36 am
Mark–Horizontal gene transfer is common in bacteria and other microbes, but a lot of researchers are of the opinion that it's not so common as to completely blur the tree-like structure of microbe phylogeny. That's why, for example, if you get an E. coli infection, your doctor will know it's an E. coli infection, and not the bubonic plague from Yersinia pestis. Horizontal gene transfer just means that species are somewhat porous among microbes. But, then again, butterflies and other animals interbreed with other species, but that doesn't mean they're not real species.
The more basic quandary about microbial species is that they don't reproduce sexually like animals. Reproductive isolation is the favorite standard for recognizing species among zoologists, but it's meaningless for bacteria. So microbiologists have come up with other ideas, such as a species being a clonal lineage that occupies a specific ecological niche, where selection weeds out mutants that aren't well-adapted to that niche.
That's hard to use in the real world to quickly identify species. A lot of scinetists use a 97% rule–ie., if the DNA of a strain of bacteria (or a section of the DNA) is more than 97% identical to a known species, then it belongs to that species. If it's less than 97% identical, it's a new species.
[(myl) I once quoted this from Betsy Dyer's A Field Guide to Bacteria:
That state of affairs — which would be more like a pattern of turbulent flow than a taxonomic tree — might still be characterized by regions with clusters of features that are worth naming; but it's not at all clear to me that this is enough to provide a coherent foundation for a question like "how many bacterial species are there?"
There seems to be a natural human urge to impose tree-structured ontologies even in areas where they don't really apply. This kind of taxonomizing is often useful, but there are some less successful episodes…]
Rosie Redfield said,
August 24, 2011 @ 11:28 am
Mark, Carl is right and Sonea is very wrong. Tree-structured ontogenies definitely apply to bacteria; the lateral gene transfer is mostly just some blurring at the edges.
[(myl) Thanks, Rosie! Can you suggest a reference that makes this argument in a more quantitative way?]
Mo said,
August 24, 2011 @ 1:22 pm
It seems to me that the whole idea of a "species" as a discrete entity is a useful fiction just like the idea of discrete "languages". I'm not sure if the number of species on earth is really a meaningful quantity.
(Interestingly I just learned the term "ring species", which appears to be the biological analogue of a dialect continuum. Interesting!)
Andy Averill said,
August 24, 2011 @ 1:46 pm
But isn't contemporary taxonomy expressed mostly in terms of genetic divergence? Which is at least theoretically quantifiable with a high degree of precision. Whereas Linnaeus and his successors relied on morphology, which is more subjective and can be misleading. Doesn't that go to the validity of these graphs? In particular, I would imagine there's been a lot of reshuffling of taxa in the years since genome sequencing became possible.
Mark Mandel said,
August 24, 2011 @ 2:17 pm
A tangential WTF?: The first figure doesn't show up at all for me in Firefox 6.0 under Snow Leopard 10.6.8. It appears in Safari 5.1, but without the hover text ("Click to barsoomenate") and barsoomenability. The second figure appears and performs right in both browsers. AFAICT the HTML is exactly parallel for both.
J. W. Brewer said,
August 24, 2011 @ 2:57 pm
The verb "barsoomenate" is a new one on me, but the first few pages of google hits seem at a quick glance to pretty much all refer to visual images of the planet sometimes known as Barsoom. What's the deal with the extended sense in the mouseover text for Mora et al.'s figure 1?
Simon Greenhill said,
August 24, 2011 @ 7:31 pm
@myl: The problem is that there's a huge debate about how prevalent LGT is in bacterial populations. It's one of the great battlefields in evolutionary biology. Some people say that it's rare and uncommon (i.e. the "just blurs the tree at the edges" comment above by Rosie Redfield), others say that it's massive (hence the quote of Sonea above).
Tal Dagan and Bill Martin's great paper The Tree of 1% is a good discussion of the problems with bacterial phylogenetics. Dagan and Martin are very clearly on the "LGT is everywhere" side of things, but present a good review of the literature and debate.
Simon
Rosie Redfield said,
August 24, 2011 @ 8:30 pm
@myl: Sonea is well out on the fringe, but there's good science on both sides of this argument. But I think the best evidence is in what's being done, not in what's being said about what should or shouldn't (or can or can't) be done.
So here's a 2009 paper from Jon Eisen's group, reporting a beautifully detailed phylogenetic tree for Bacteria and Archaea: A phylogeny-driven genomic encyclopedia of Bacteria and Archaea.
Steve Morrison said,
August 24, 2011 @ 8:31 pm
I believe the verb "barsoomenate" was coined by Phil Plait of the Bad Astronomy blog as one of his numerous synonyms for "embiggen"; however, I'm also puzzled by its appearance in this context!
Jair said,
August 25, 2011 @ 1:53 am
There is an excellent article by Stephen Jay Gould, "Species are not specious", about the question of to what extent species are socially constructed rather than objective. You can probably guess his opinion by the title. It can be found in Google Books. I don't believe it discusses the issue of microbial species, however.
Mr Punch said,
August 25, 2011 @ 1:30 pm
Was there really a G.H. Toulmin in 1956? Or is this somehow a "cover" for Turing, who had died in disgrace not long before? The only G.H. Toulmin of whom I'm aware wrote in the late 18th century on the antiquity of the earth (pretty old, he reckoned).
A noble heart embiggens the smallest planet – Telegraph Blogs said,
August 28, 2012 @ 10:12 am
[…] Mars post has next to it: "Click to barsoomenate". That one was new to me, but I've Googled it and found it used in the context of "display larger version of this picture" on as weighty an authority …, so I think that's a neologism that's sticking. I think it works because the middle syllable sounds […]
A noble heart embiggens the smallest planet | Alkaon Network said,
August 28, 2012 @ 11:16 am
[…] to it: "Click to barsoomenate". That one was new to me, but I've Googled it and found it used in the context of "display larger version of this picture" on as weighty a…, so I think that's a neologism that's sticking. I think it works because the middle […]