Divergent histories of languages and genes
« previous post | next post »
Charles Darwin saw the history of languages as a model for "descent with modification" in biological evolution; and researchers from Thomas Jefferson to Luigi Luca Cavalli-Sforza and beyond have been excited about the idea of combining linguistic, biological, and geographical evidence to shed light on the history of human populations.
Most recent linguists and anthropologists who are knowledgeable about such topics have been skeptical about how close we should expect linguistic and biological descent to be, in general. There are many ways, both wholesale and retail, for people to end up speaking a language different from the language of their ancestors, and similarly many ways for genes to flow from one speech community to another.
A recent contribution to the skeptical side of the discussion is Hafid Laayouni et al., "A genome-wide survey does not show the genetic distinctiveness of Basques", Human Genetics, published online 1/16/2010.
Here's their abstract:
Basques are a cultural isolate, and, according to mainly allele frequencies of classical polymorphisms, also a genetic isolate. We investigated the differentiation of Spanish Basques from the rest of Iberian populations by means of a dense, genome-wide SNP array. We found that FST distances between Spanish Basques and other populations were similar to those between pairs of non-Basque populations. The same result is found in a PCA of individuals, showing a general distinction between Iberians and other South Europeans independently of being Basques. Pathogen-mediated natural selection may be responsible for the high differentiation previously reported for Basques at very specific genes such as ABO, RH, and HLA. Thus, Basques cannot be considered a genetic outlier under a general genome scope and interpretations on their origin may have to be revised.
There's an excellent discussion of the article by Razib Khan, "The Basques may not be who we think they are", Gene Expression 2/18/2010.
(I believe that "FST" is a form of Wright's coefficient of relatedness — see e.g. Hilde M. Wilkinson-Herbots, "Coalescence Times and FST Values in Subdivided Populations with Symmetric Structure", Advances in Applied Probability 35(3) 2003. I guess I should point out that the analysis in the Laayouni paper is based on a linear (SVD) decomposition of (a relatedness measure derived from) a large (~280k) number of SNP frequencies. I don't know enough about the methods and the assumptions behind them to evaluate how strong an argument their results constitute against "the genetic distinctiveness of Basques" — I suspect that the phrase chosen in their title, "…does not show the genetic distinctiveness of Basques", is a fair way to put it.)
For some general linguistic background, see Don Ringe, "The linguistic diversity of aboriginal Europe", 1/6/2009.
Tim Silverman said,
February 21, 2010 @ 8:41 am
It is a bit weird the way most of Khan's article (I guess reflecting its sources) assumes "non-Indo-European" = "pre-Neolithic", given that the people who spread the Neolithic (almost certainly) weren't Indo-European-speakers. He even says this at the end, but it would have been better earlier.
marie-lucie said,
February 21, 2010 @ 2:57 pm
These studies seem to assume that the Basques did not intermarry with other populations. Intermarriage in the geographical zones close to the present-day Basque country must have occurred: for instance in Gascogne, previously written Guascogne, from the evolution of the Latin name Vasconia, where the root vasc- ([wask]) correspond to present-day "Basque" (which is a French spelling for a Spanish-influenced pronunciation). The typical Basque appearance (short, wiry, dark-haired, with sharp features) is also common throughout the adjoining regions.
Trond Engen said,
February 21, 2010 @ 3:20 pm
This is as one should expect, given the long coexistence of Basque and non-Basque speakers in the region. Genes, languages and culture drift across the map at different paces and driven by different forces. Where I believe that genes, or archaeology, can throw light on linguistic issues, is at events of transition, as indications of the sort of major upheaval that might have triggered a massive language shift. Or vice versa: A known language shift may throw light on some archaeological or genetic event.
Similarly the genetic variation within Saami and non-Saami populations in Scandinavia is dwarfing any difference between them. But still, somewhat diconcerting: As marie-lucie can distinguish a typical Basque appearance, so can I distinguish a typical Saami. I suppose the point is that the collocation of "typical" (visible or invisible genetic) traits with certain cultures and/or languages is a matter of course: Both have geographical distributions and some are bound to coincide at any time. Give or take a millennium or five and the alignment is totally different.
John Cowan said,
February 21, 2010 @ 5:37 pm
The Xhosa alone have 80% of humanity's genetic diversity.
[(myl) It's hard to know what that would mean for this kind of test, though. At one extreme, the Xhosa could exhibit a lot of group-internal SNP diversity that is mostly not shared with others. At the other extreme, all variant SNPs found anywhere would also be found among the Xhosa. I expect that the truth is somewhere in between — and probably depends on what set of SNPs you look at, and how you weight them. The point of this paper is that (in their analysis) Spanish Basques look like other Spaniards, and French Basques look like other people in non-Iberian Europe, and whatever the various subgroups of Basques have in common is also shared with non-Basques:
]
John Cowan said,
February 21, 2010 @ 6:34 pm
I meant the latter: within-group variation swamps between-group variation.
Carl Anderson said,
February 22, 2010 @ 10:59 am
I think the points about how there has obviously been a great deal of intermarriage between Basque-speakers and non-Basque-speakers over the years have already been made in some comments above — but what confuses me is why this isn't obvious, and why it is still possible to act like it's news that Basque-speakers _aren't_ a genetic isolate? What gave anyone the idea that they _should_ be?
[(myl) There's a fair amount of "folk genetics", perhaps especially among Basques themselves, that assumes this idea. And the start of the Laayouni et al. paper explains that there are some previous experimental results supporting this view:
Those references are to Chalmers JN, Ikin EW, Mourant AE (1949) The ABO, MN and Rh blood groups of the Basque people. Am J Phys Anthropol 7:529–544; Bertranpetit J, Cavalli-Sforza LL (1991) A genetic reconstruction of the history of the population of the Iberian Peninsula. Ann Hum Genet 55:51–67; Calafell F, Bertranpetit J (1994a) Mountains and genes: population history of the Pyrenees. Hum Biol 66:823–842.
The conclusion of Laayouni et al. is that the polymorphisms for which the Basques have distributions that are different from their neighbors (mainly blood-type differences) are not a residue of genetic origins, but rather a "microgeographic" adaptation:
]
After all, even a cursory examination of Spanish reveals a fair amount of Basque has gotten into the daily vocabulary, such that it is difficult to imagine so much linguistic exchange with people, well, making a bunch of babies over the past 2000 years. Moreover, presumably Romance languages weren't spoken in the Iberian penninsula until Latin-speakers showed up, which implies that every Romance-speaker in Iberia today probably has a few pre-Roman-era ancestors who spoke _something_ non-Romance, possibly including earlier forms of Vasconic … so unless the act of speaking a Romance language mutated everyone's genes, it's difficult to imagine why we should assume modern Basque speakers should be very distinct, genetically, from modern non-Basque-speaking inhabitants of the Iberian penninsula (or southwestern France, for that matter, I guess).
Trey said,
February 22, 2010 @ 11:33 am
Here's an example of why I don't trust anyone who is terribly enthusiastic about combining biological genetics and historical linguistics. This is an Italian translation (all I could find online) of a diagram made by Cavalli-Sforza and Merrit Ruhlen.
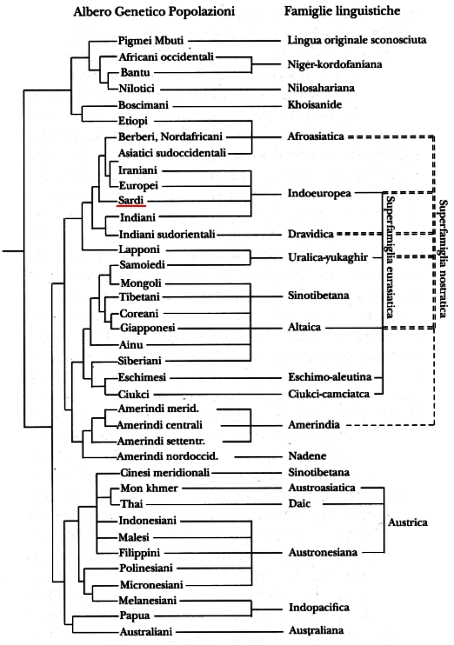
This diagram lets casual observers infer that there is an incredibly strong correlation between the genetic classification and the linguistic classification because the leaf nodes of the two trees can be aligned. The order of the leaf nodes is completely irrelevant—what matters is the structure of the tree. And the structures fail to line up in very significant ways.
Take Ethiopic (Etiopi) and Berber (Berberi). On the linguistic side, they are as close as they can be—they are siblings under Afroasiatic. On the genetic side, they are as far apart as they can be, structurally—their lowest common ancestor is the root of the entire tree. Similarly, Ethiopic and "Boscimani" are siblings on the genetic side, but are unrelated on the linguistic side, even though Ethiopic goes all the way up to the controversial Nostratic super-family on the linguistic side, it still can't hook up with "Boscimani"'s Khoisan linguistic roots. There's a similar triple with Lappish (Lapponi), Finnish (Samoiedi) and Mongolian (Mongoli). Lappish and Finnish are linguistic siblings, but the Lappish are more closely related, genetically, to the Berbers than to the Finns, and the Finns are more closely related, genetically, to the American Indians than to the Lappish. And sticking the unattached Sino-Tibetan sub-tree under, but unattached to the Eurasiatic super-family is just a trick to make the linear alignment of the leaf nodes come out right—and Sino-Tibetan actually got broken in two (look for Cinesi meridionali in the bottom third of the chart) to make it fit the genetic classification.
What I see is the exact opposite of what Ruhlen used this diagram to claim (I reviewed his book with this chart in it back in the 90's)—there's a rough alignment in many places between the linguistic and genetic trees because for most of human history, most humans taught mostly their own language to children who mostly their own. And I bet most adoptions were local, and within the same ethnic group, which wouldn't complicate the picture. But as you said, there are many ways for people to end up speaking a language different from the language of their ancestors.
Lecturas biolingüísticas « Biolingüística said,
February 22, 2010 @ 11:48 am
[…] Como es sabido, la lengua vasca es una cosa un poco rara en la distribución lingüística europea: no es una lengua romance, no es una lengua sajona. Se supone que es una lengua pre románica autóctona. Algunos incluso han dicho que podría tratarse de una lengua que proviene de los primeros grupos humanos en la zona. En términos genéticos, esto conllevaría en cierta forma a que los vascos posean (por decirlo mal y pronto) alelos más viejos que los del resto de los europeos. Pues bien, un estudio reciente titulado A genome-wide survey does not show the genetic distinctiveness of Basques demuestra que esto no es cierto, que los vascos tienen la misma diferencia genética con otro europeo que un griego, un suizo o un italiano. El artículo ha sido comentado en Language Log (click aquí). […]
Mary Kuhner said,
February 22, 2010 @ 6:21 pm
It is an ongoing problem in phylogenetics, including linguistic applications, that the node order of a tree has no scientific meaning but makes a HUGE difference in how readers will interpret the tree. I've seen some pretty blatant exploitation of this effect in the human genetics literature.
Even worse, many genetic trees contain ties, and tree-makers often silently resolve those ties in a way that favors their hypothesis. I read one paper in which almost all of the evidence for correlation between geography and genes came from using geography to break ties in the gene tree. (The actual data were too scanty to make a well-resolved tree, but if this had been acknowledged the paper would probably not have gotten published.)
It's not clear how to fix this. The tree has to be shown with some node order or other, but showing it with a node order chosen to support your hypothesis can be extremely misleading. Requiring authors to alphabetize their samples just strikes me as encouraging manipulation of the (usually arbitrary) sample names. Requiring a random order is hard to enforce. ("Sure, it came out this way randomly! Can you prove it didn't?")
I make my undergrads do problems about this issue, and they seem to get the point, but will they remember it years later if they aren't doing phylogenetics themselves? And what about lay readers?
Nathan Myers said,
February 22, 2010 @ 7:06 pm
Two observations struck me about this revelation. First, astonishment that Iberians are so reliably distinct from other western Europeans. (Does the Moorish admixture show? Are all Iberians genetically Basques?) Second, the insistence that events of the past two or four thousand years would be presumed to trump those of the twenty thousand years previous. Surely wave after wave of migrations, displacements, and genocides swept through unrecorded, leaving Europe unreadably scoured and littered by their passage. Any order that can today be discerned ought first to be proven not accidental before using it to support a claim. I don't know any way to prove such a thing.
[(myl) The cited paper suggests that adaptation to local conditions (for example, endemic diseases) is an important factor. This can create geographical variation, independent of the residue of migrations (which surely does also exist). It's also important to note that what patterns you see apparently also depends on what markers of genomic variation you look at, and probably also on what algorithms you use to infer the patterns.]
Does the late demonstration by Carl Woese that horizontal gene transfer dominated evolution during most of life's history, only recently supplanted by Darwinian natural selection in latecomers like us, hint that its parallel with linguistics, the loan word, may be more important than was thought?
[(myl) It's not really a matter of calibrating importance. The key thing is that neogrammarian-style "sound laws" (and some other systematic changes) are common, and leave a residue that can be used to reconstruct (aspects of) earlier linguistic stages (including often a time-sequence of stages), demonstrate historical relationships, place borrowings in relative time, and so on. This remains true even when large portions of a language's vocabulary are borrowed (as is often the case).
The point is not that linguistic history is mostly a matter of inheritance with mutation-like modifications, as opposed to horizontal-transfer-like borrowing. The point is that there's a technique for inferring historical relationships based on analyzing the residue of the "mutations", which also helps to identify and date the "horizontal transfers".]
Ken Brown said,
February 23, 2010 @ 10:41 am
> The conclusion of Laayouni et al. is that the
> polymorphisms for which the Basques have
> distributions that are different from their
> neighbors (mainly blood-type differences) are
> not a residue of genetic origins, but rather a
> "microgeographic" adaptation:
[…]
>> The contradiction with previous
>> reports that depicted Basques as genetic
>> outliers can be resolved if we consider that
>> the polymorphisms accounting for most of this
>> differentiation lie in genes such as ABO, RH,
>> and the HLA complex that are, given their
>> involvement in host–pathogen interactions,
>> obvious targets for natural selection in the
>> ancestral populations even at a
>> microgeographic scale.
This might be the biologically most interesting observation. It implies that we might be able to use this sort of evidence to show natural selection creating sub-populations distinguishable by those few genes, even though there is a large amount of interbreeding between them and other genes flow easily between the them.
Good evidence of natural selection is always fun. But it also supports the idea that the structure we see in human populations depends on which genes we look at. So you could construct many different "races" by paying attention to different sets of genes. Dividing the world up on the basis of facial appearance gets us one structure, if we use skin colour another, blood groups another, and any small set of genes would give us different subgroups that probably wouldn't correlate well with each other. Which is consistent with our current model of very rapid expansion from a small(ish) group of founders and no long-term genetic isolation between major human sub-populations, but does not fit with popular ideas of ancestral populations splitting into mutually exclusive races that could be diagrammed as a tree.
Or in cladistic jargon, humans really are connected by tokogenetic webs, not phylogenetic trees. The methods of phylogeny do not describie relationships within a species very well, other than for single genes. Which geneticists knew all along I suppose, but all those trees they draw of things like Y-chromosomes or mDNA are often interpreted as if they represented migrations of distinct clans or tribes or races – but they really don't and they really can't.
So even if trees are appropriate models for the development of language, there can be no relevant genetic trees to compare them with. (You can look at clusters rather than trees, but that's not the same thing)
Josh said,
February 23, 2010 @ 11:40 am
Trey, could many of the mismatches between languages and genes be due to the fact that Cavalli-Sforza used classical genetic markers to determine relationships between populations? As Razib points out in his post, in the 1990s Cavalli-Sforza claimed, drawing on classical markers, that the Basques were a genetic isolate, whereas more modern methods that look into uniparental markers or large numbers of autosomal markers conclude that they are not different from other Europeans.
Regarding classical markers, Razib says that "there's now evidence that blood group distributions are not random, and may emerge as responses to disease pressures. In other words, they aren't neutral markers which give a good sense of ancestry".
This recent editorial in Current Biology by Colin Renfrew discusses the possibility of a synthesis of linguistics, population genetics, and archaeology: "Archaeogenetics — Towards a ‘New Synthesis’?"
Trey said,
February 23, 2010 @ 3:44 pm
Josh, I don't know what effect the specific genetic markers tested have on the value of the results. But my major concern isn't the quality of the biological-relatedness tree (for the moment I'll trust the biologist to do the biologist's work), it's the misleading presentation of the results, which gives the casual observer the impression that the trees are in near perfect alignment. Minor mis-alignments, or minimally shuffling the order of small subtrees to make the graph look pretty is one thing, but the treatment of Sino-Tibetan, for example, seems to be intentionally misleading. A simple line connecting the two occurrences of it would make it clear that there were two occurrences, but then the linguistic tree would obviously be folded on itself.
Despite my earlier strong words, I'm not wholly against the use of linguistic data alongside genetic, historical, archeological, and other information to figure out how people and cultures are related. It's a very interesting area of study. The mention of Cavalli-Sforza (and by association the thought Merrit Ruhlen) just sets me off because it reminds me of things like this artificial tree alignment, presented without all the necessary caveats for the non-tree-savvy readers. (And in the book I first saw it in, it was alongside Ruhlen's horrible mass comparisons and superfamily etymologies—some of them even made my wife (who is not a linguist) cringe and ask, "really? can they do that?")
I expect any scientist or researcher doing comparison of cladistic hierarchies (biologist or linguist) to be tree-savvy in a good way, not using their powers for evil, misleading readers who may not have time, interest, or aptitude to analyze the tree carefully to find structural anomalies, while maintaining plausible deniability should someone be so rude as to point them out.
Ingrid Jakobsen said,
February 24, 2010 @ 12:52 am
And since no-one else seems to have said this, FST is the biggie of the F statistics developed by Sewall Wright, and has more different definitions than anything else I can think of in population genetics.
» On the autochthony of the Basques CQ2 | Ed Murphy said,
February 24, 2010 @ 9:10 am
[…] the always-worth-reading Language Log, an article entitled "A genome-wide survey does not show the genetic distinctiveness of […]
Jubin said,
February 25, 2010 @ 1:57 am
My family is 3/4 Iranian Kurdish, 1/4 Basque.
Not only is it hard to tell the difference between the Basque side of my family and most other Spaniards; it's hard to tell the Basque side of the family from the Kurdish side.
Many phenotypical genetic differences are more or less arbitrary, in this case, probably have more to do with mountains than anything.
Mike said,
July 23, 2010 @ 9:04 pm
First of all, as many have pointed out…a modern population can be descended from several ancient populations. And it is true, even one individual carrying genes from one population into another, every generation, can create enough genetic admixture to counter the genetic drift that, over time, will differentiate populations that have zero gene flow between them. So it is not surprising that the Basque are genetically similar to their neighbors. However, just because modern population X is not distinct does not rule out the possibility that the ancestors of many individuals in population X were from a distinct, or ancient, group.
The fact that geography influences and constrains the degree of genetic relatedness was wonderfully illustrated by John Novembre et al. in "Genes mirror geography within Europe", Nature 456,98-101 (2008).
You can see that if one rotates the coordinates in their PCA plot of over 500,000 SNPs, you get a fairly good representation of Europe! And the Basque individuals are where you'd expect them to be…clustered with other Iberians. However, when you look at maternal ancestry with mtDNA, although there is no sharp demarcation between Basques and neighbors, there is a gradient of increasing frequencies of haplogroups H1 and H3, (see it in Achilli et al. "The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool", Am. J. Hum. Gen. 75:910-918, 2004). That to me suggests that the Basques are smack dab in the middle of where an ancient demographic expansion happened…so many modern Basques are surely descended from those probably pre-agricultural populations. Many, but not all. You just can't talk about populations as if they were individuals…they are semi-permeable.
Anyway, thats my 2 cents.
Mike said,
July 23, 2010 @ 9:10 pm
Sorry, I mis-spoke. Novembre's article did not have a sample that was specifically "Basque". But if you consider the recent papers, you would not expect the Basques to cluster far from the Iberian "cloud" in principal coordinate-space.
Also, I didn't explain the gene flow thing very well. What I meant was, IF no gene flow occurs, over time (because of random changes in allele frequency call genetic drift) two separate populations tend to become more and more genetically "distinct" (even if they were originally ONE population!). But with a little gene flow…they don't become very distinct…there will be a relatively low Fst value for them. Relatively low structure.