Mice with the "language gene" stay mum
« previous post | next post »
 Now they've done it — spliced human FOXP2, often called the "language gene", into some mice in Leipzig. This won't give the mice anything new to say, but many people were certainly expecting them to start producing and analyzing more complex sound patterns. Thus Juan Uriagereka ("The Evolution of Language", Seed Magazine, 9/25/2007):
Now they've done it — spliced human FOXP2, often called the "language gene", into some mice in Leipzig. This won't give the mice anything new to say, but many people were certainly expecting them to start producing and analyzing more complex sound patterns. Thus Juan Uriagereka ("The Evolution of Language", Seed Magazine, 9/25/2007):
Chimps, and our other close relatives the apes, certainly have the hardware for some basic forms of meaning […]. What they don’t have is a way to externalize their thoughts. I’d wager that chimps just lack the parser that FoxP2 regulates.
Uriagereka suggested that "Because of the similarities in brain structure and in the syntax of their song, finches must also have this parser", created by the songbird version of Foxp2. If this bold conjecture were true — that certain alleles of this particular gene create a "parser" in the brain — then the mice recently gifted with a "humanized" form of foxp2 should exhibit some striking abilities, such as recursively-structured squeaks.
In fact, of course, this didn't happen. By the breathless standards of mass-media talking-animal stories, the results were underwhelming — and today in the NYT, Mark Leyner's Op-Ed ("Of Mice and Monologues") comments on the resulting lack of media uptake:
I AM absolutely baffled as to why the announcement of a scientific advance heralding the advent of talking mice has not generated a peep from the chattering classes, particularly since it’s a story about chattering … and chattering mice, to boot.
But what did happen is still very interesting, and probably does tell us something about the evolution of language. And just as interesting, this story offers an object lesson in the limitations of the "gene for X" idea: the notion that specific behavioral abilities — or for that matter specific anatomical structures or physiological processes — are typically associated with single genetic variations.
The new work is documented in a recently published paper, Wolfgang Enard et al., "A Humanized Version of Foxp2 Affects Cortico-Basal Ganglia Circuits in Mice", Cell, available online 5/28/2009. The same journal issue also includes an editorial discussion by Philip Lieberman, "FOXP2 and Human Cognition".
But before describing what Enard et al. did to those mice, and what happened as a result, and why it wasn't splashed all over the media, let's do some scientific due diligence, in the form of a review of the factual background.
The gene in question is "Forkhead box P2", one of many genes for "Forkhead box" proteins, a large family of transcription factors. The forkhead box part refers to a DNA-binding structure consisting of about 100 amino acids in a characteristic pattern, found in 17 known families of proteins denoted by added letters from FoxA to FoxQ. So the name is composed of fox="protein with the forkhead box structure", P="the 16th type of fox protein", and 2="the second subtype of FoxP protein". A version of FoxP2 is found is fish, reptiles, and birds as well as mammals.
Wolfgang Enard et al. "Molecular evolution of FOXP2, a gene involved in speech and language", Nature 418: 869-872, 2002, sequenced the DNAs that encode the FOXP2 protein in chimpanzee, gorilla, orang-utan, rhesus macaque and mouse. They reported surprisingly small differences:
FOXP2 (forkhead box P2) is located on human chromosome 7q31, and its major splice form encodes a protein of 715 amino acids belonging to the forkhead class of transcription factors. It contains a glutamine-rich region consisting of two adjacent polyglutamine tracts, encoded by mixtures of CAG and CAA repeats. Such repeats are known to have elevated mutation rates. In the case of FOXP2, the lengths of the polyglutamine stretches differed for all taxa studied. Variation in the second polyglutamine tract has been observed in a small family affected with speech and language impairment, but this did not co-segregate with disorder, suggesting that minor changes in length may not significantly alter the function of the protein. If the polyglutamine stretches are disregarded, the human FOXP2 protein differs at only three amino-acid positions from its orthologue in the mouse. When compared with a collection of 1,880 human–rodent gene pairs, FOXP2 is among the 5% most-conserved proteins. The chimpanzee, gorilla and rhesus macaque FOXP2 proteins are all identical to each other and carry only one difference from the mouse and two differences from the human protein, whereas the orang-utan carries two differences from the mouse and three from humans. [emphasis added]
Why so much interest in this particular gene, out of the tens of thousands available to study? Back on the 1990s, several members of a British family, over several generations, were found to share a syndrome whose symptoms include "defects in processing words according to grammatical rules; understanding of more complex sentence structure such as sentences with embedded relative clauses; inability to form intelligible speech; defects in the ability to move the mouth and face not associated with speaking (relative immobility of the lower face and mouth, particularly the upper lip); and significantly reduced IQ in the affected compared with the unaffected in both the verbal and the non-verbal domain". (The quoted desription comes from an excellent 2003 summary by Alex McAndrew.)
Genetic studies showed that the affected family members had a mutation involving two amino-acid substitutions in a single gene on chromosome 7 (7q31), associated with several abnormalities in brain structure and function. McAndrew again:
The most significant abnormality was bilateral reduction in the size of caudate nucleus (a component of the basal ganglia) coupled with abnormal high activity in the left caudate nucleus during speech tasks. The caudate nucleus is implicated in motor co-ordination and also processes information that is being sent from other areas of the brain to the frontal lobe. Broca's area, important for speech production, was also smaller and over-activated during speech production in affected subjects.
Given the natural but unfortunate human propensity to try to line effects up one-to-one with causes — in this domain, the "gene for X" syndrome — it's not surprising that FOXP2 has often been called "the language gene".
We've discussed FOXP2 a number of times over the years — a good place to find an overview is Geoff Pullum's post "The continuing misrepresentation of FOXP2 effects", 9/5/2005, which cites McAndrew at greater length. News since 2003 includes a 2004 study of foxp2 expression in songbirds, a 2005 study of mice with disrupted foxp2, a 2005 study of FOXP2 truncation in human cases of verbal dyspraxia, and a 2007 study of foxp2 evolution in echo-locating bats.
In mice, disrupting ("knocking out") both copies of FOXP2 results in "severe motor impairment, premature death, and an absence of ultrasonic vocalizations that are elicited when pups are removed from their mothers". Disrupting one copy resulted in "modest developmental delay but a significant alteration in ultrasonic vocalization". The knockout mice had several abnormalities in brain development, incluing less elaborate dentritic arbors in cerebellar Purkinje cells.
Some additional information and links can be found in the Wikipedia article on FOXP2. [A typographical note: human genes are generally spelled with all capital letters, while mouse and other animal genes are generally spelled with just an initial capital letter, or with all lower case letters. So the spelling of the human gene (and protein) in this case is always "FOXP2", while the animal genes (and proteins) are sometimes "Foxp2", sometimes "FoxP2", and sometimes "foxp2".]
In the new study, Enard et al. looked at the effects of introducing some aspects of normal human FOXP2 into mice:
It has been proposed that two amino acid substitutions in the transcription factor FOXP2 have been positively selected during human evolution due to effects on aspects of speech and language. Here, we introduce these substitutions into the endogenous Foxp2 gene of mice.
What they actually did is a bit complicated, but worth understanding in detail:
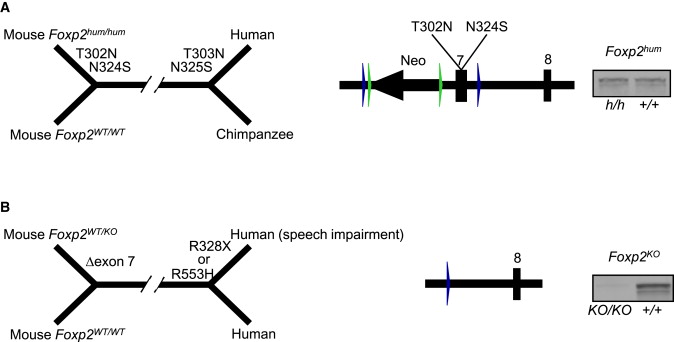
Individuals that are heterozygous for FOXP2 alleles that carry a missense mutation (R553H) affecting the forkhead DNA binding domain of the protein, a nonsense mutation (R328X) or disruptions of the gene by a chromosomal rearrangement suffer from a developmental impairment especially affecting speech and language […] Analyses of the evolution of the FOXP2 gene in primates identified two amino acid substitutions (T303N, N325S), which became fixed on the human lineage after its separation from the chimpanzee and which appear to have been subject to positive selection […].. It has been hypothesized that these substitutions underwent selection due to effects on some aspects of speech and language […]. Conveniently, FoxP2 in chimpanzees differs from Foxp2 in mice by only one conservative amino acid substitution (D80E). Thus, the wild-type mouse Foxp2 protein can be regarded as a model for the ancestral version of the human FOXP2 protein and compared to a partly “humanized” version in which the two amino acid replacements have been introduced.
Thus the mice with "humanized" Foxp2 (or should it be FOXP2?) actually have a gene that still differs in one location (those pesky glutamine tracts aside) from the human type.
So what happened?
Although these mice are generally healthy, they have qualitatively different ultrasonic vocalizations, decreased exploratory behavior and decreased dopamine concentrations in the brain suggesting that the humanized Foxp2 allele affects basal ganglia. In the striatum, a part of the basal ganglia affected in humans with a speech deficit due to a nonfunctional FOXP2 allele, we find that medium spiny neurons have increased dendrite lengths and increased synaptic plasticity. Since mice carrying one nonfunctional Foxp2 allele show opposite effects, this suggests that alterations in cortico-basal ganglia circuits might have been important for the evolution of speech and language in humans.
Those "mice carrying one nonfunctional Foxp2 allele" are the knock-out mice, with one normal mouse Foxp2 and one knocked out — the mice with two non-functional Foxp2 alleles, you'll recall, basically just died.
Did the humanized-FOXP2 mice produce calls that were more complex, more (so to speak) "parsed" or at least parsable than their wild-type ("wt") littermates? Well, no — the human gene caused a small change, in the direction of a certain type of call being lower in pitch and "less modulated":
Finally, to assess whether Foxp2hum impacts vocalization, we recorded ultrasonic vocalizations emitted by pups when placed outside the nest at day P4, P7, P10, and P13 from 32 Foxp2hum/hum mice and 39 Foxp2wt/wt littermates. Using a semi-automated procedure to extract single calls and a general linear model (GLM) with the variables postnatal day, genotype, sex, litter and weight we found no significant differences in the number of calls emitted per minute or in the duration of intervals between calls. To analyze the structure of calls, we assigned them to one of four categories: (1) calls shorter than 50 ms with no frequency jumps; (2) calls longer than 50 ms with no frequency jumps; (3) calls with frequency jumps; and (4) remaining sounds, which were not analyzed. The first call type, which was the most frequent, showed no difference between the two genotypes with regard to the number of calls, the duration of calls and five other parameters. However, Foxp2hum/hum animals, they had a significantly lower start peak frequency (p < 0.001), and lower mean (p < 0.01), minimum (p < 0.01) and maximum (p < 0.001) peak frequencies. In addition, the slope of the calls declined less in frequency (p < 0.01, Figure 6B) and were locally less modulated (p < 0.01). […]
[C]alls with frequency jumps lasted longer ( p < 0.05), had longer gaps (p < 0.05) and started (p < 0.01) and ended (p < 0.05) with higher peak frequencies in Foxp2hum/hum mice than in their wild-type littermates.
Enard et al. stress that "it is important to note that this influence [on vocalizations] is subtle and within the range of normal variation among mice". So these mice with the human "language gene" didn't chatter — they just squeaked a little more gruffly and plainly than their wild-type littermates in their short and continuous calls, and squeaked a little higher and more slowly in their calls with frequency discontinuities.
That doesn't mean that the Enard et al. results were entirely negative. Those "increased dendrite lengths" and that "increased synaptic plasticity" in the striatum are very interesting and suggestive, since such changes might plausibly make important differences in a circuit involving the frontal cortex and basal ganglia that is known to play a role in speech and language.
The general idea that those brain regions play an important part in speech and language is not controversial, though there are several different ideas about the nature of the role that they play. One interesting perspective is the "Declarative/Procedural Model", which suggests that "mental grammar involves procedural memory and is rooted in the frontal cortex and basal ganglia" (see Michael Ullman, "A Neurocognitive Perspective on Language: The Declarative/Procedural Model", Nature Reviews Neuroscience, 2:717-726, 2001). I find this idea much more plausible than the Piattelli-Palmarini/Uriagereka idea of a "parser" in the caudate nucleus, created by a two-amino-acid change in FOXP2.
An alternative (and essentially non-linguistic) interpretation of (much of) the same evidence is offered by J.M. Novick et al., "Cognitive control and parsing: Reexamining the role of Broca's area in sentence comprehension", Cognitive, Affective, & Behavioral Neuroscience, 5: 263-281, 2005. They suggest that
(1) [Left Inferior Frontal Gyrus] is part of a network of frontal lobe subsystems that are generally responsible for the detection and resolution of incompatible stimulus representations; (2) the role of LIFG in sentence comprehension is to implement reanalysis in the face of misinterpretation; and (3) individual differences in cognitive control abilities in nonsyntactic tasks predict correlated variation in sentence-processing abilities pertaining to the recovery from misinterpretation.
Novick et al. also note that "representational conflict can arise at numerous possible stages, ranging from internal perceptual representations all the way to conflicting motor-response preparations".
I'm fond of the idea that syntactic structure, both in production and in perception, is a sort of evolutionary overlay on a system originally developed for learning and executing complex motor patterns. Working out the neural architecture of motor control that underlies such patterns — linguistic and otherwise — is one of the most exciting research topics around these days, in my opinion. But even if this idea is correct, the evolutionary pathways surely involve the interaction of many genes affecting structure and function. And the contribution of each one is probably something like an increase in the capacity of certain brain subsystems — like those "increased dendrite lengths" and "increased synaptic plasticity" in the striatum — rather than the miraculous genetic pre-programming of a neural parser in the caudate nucleus by the substitution of two amino acids in one gene.
Returning to the problem of media uptake, Mr. Leyner is not quite correct that "the announcement of a scientific advance heralding the advent of talking mice has not generated a peep from the chattering classes". Nicholas Wade is certainly a member in good standing of that set, and he dutifully covered the Enard et al. paper: "Human Language Gene Changes How Mice Squeak", NYT 5/28/2009. And he did his best with what came to hand:
Svante Paabo, in whose laboratory the mouse was engineered, promised several years ago that when the project was completed, “We will speak to the mouse.”
He did not promise that the mouse would say anything in reply, doubtless because a great many genes must have undergone evolutionary change to endow people with the faculty of language, and the new mouse was gaining only one of them. So it is perhaps surprising that possession of the human version of FOXP2 does in fact change the sounds that mice use to communicate with other mice, as well as other aspects of brain function.
Reporters don't write headlines, and so Wade is not responsible for the "human language gene" phrase in the headline. And he does end with a quote from Gary Marcus: “People shouldn’t think of this as the one language gene but as part of a broader cascade of genes,” he said. “It would have been truly spectacular if they had wound up with a talking mouse.”
Indeed. Truly spectacular, and also completely and totally unexpected — unless you've been reading, and believing, the many stories over the years that use the phrase "language gene" to describe FOXP2. The overselling of this idea originates with (certain) scientists, alas. But journalists like Wade have aided and abetted them, probably because the "language gene" idea makes a much better story than the more complex alternatives, and has only the incidental and unimportant handicap of being false.
[The cartoon panel at the top of this post is from Stephan Pastis' Pearls Before Swine for 5/20/2009.]
[Update — just to see the extent to which smart people can mislead themselves about such things, see the column "Talking Mice May Happen Before Pixar's Translated Dog", AMC SciFiScanner, which links to the article in Cell, and describes it thus:
[T]he Max Planck Institute of Leipzig published in Cell Magazine the results of their experiments breeding a group of mice with a human Foxp2 implanted in their basal ganglia (the speech center of the brain). According to their findings, the speechified mice had a better capacity for language, and "talked" with a low pitched whistle rather than their usual mousey squeak.
I suspect we can expect to see talking mice in the movies before long. ]
Sili said,
June 4, 2009 @ 1:45 pm
Ah. But did the transgenic mice have bigger crockuses? (crocki?)
carla said,
June 4, 2009 @ 1:50 pm
No substantive comment here – I just wanted to thank Prof. Liberman for providing such a thorough, and thoroughly interesting, background on the molecular biology behind the experiment.
Faldone said,
June 4, 2009 @ 2:08 pm
Anybody knows that mice can already talk. Anybody, that is, who knows that humans are not the second most intelligent species on the planet, but the third most intelligent.
Boris said,
June 4, 2009 @ 2:47 pm
Wow, had I not seen the movie version a couple of weeks ago, I would have no idea what you're talking about!
Faldone said,
June 4, 2009 @ 2:57 pm
Most people just smile and nod.
Tim Silverman said,
June 4, 2009 @ 3:04 pm
Well, thinking that taking a gene for a transcription factor from humans and transferring it to mice will cause mice to acquire syntax is about like thinking that if you take the on-off switch out of your desk lamp and put it in your radio, it will cause your radio to light up.
But even within the world of the sane, this seems like a singularly unenlightening experiment. Basically, if you take the mechanism that builds a part of the brain that's responsible for vocalisation and randomly bugger it up, then that damages the animal's ability to vocalise. That's really not a surprise. I don't really see what is to be gained by this sort of thing. It would make sense to look at what the direct effect of fiddling with transcription factors is on the regulatory networks that they're involved in, but to do that, you have to understand the regulatory network in the first place. (Besides which, the differences between humans and mice with respect to FoxP2 presumably involve not just the gene itself but also its targets. There's no obvious reason to believe that FoxP2 changed first.)
It would also make sense to relate changes in the gene regulatory network to changes in brain structure, but to do that we'd need to have a better idea of how brain structures get built, in some detail. I'd much rather see work elucidating these details. And to understand how changes in genes affect function, we'd also need to understand much better how the brain structures are responsible for function. All the actual knowledge lies in these nitty-gritty details. Experiments which say, basically, "If you randomly bash things with a stick, they break," don't add so much. If this work sets off a train of research that gets at the cascading effects of mutations in the network to which FoxP2 belongs, then that would be a very good thing. But by itself, it tells us very little. (But of course this is true of almost all research results—it's not to be held against the experiment itself.)
(Hmm, I seem less annoyed at the end of my comment than I was at the beginning. I guess that is progress of sorts.)
[(myl) Well, if you remove all the "language gene" discussion, then what they did was to make a two-amino-acid change in one gene, and what happened (among some other things) was an increase in dendritic arborization in a certain brain-stem structure. That's one of those "nitty-gritty details", right?
Now, without the "language gene" associations, Nicholas Wade never would have written up the experiment, and for that matter, the 56 (!) authors probably never would have done the experiment in the first place. But all the same…
And like in the joke about the thief and the king, who knows, the
horse might learn to singmice might learn to talk! ]mgh said,
June 4, 2009 @ 4:38 pm
Tim,
I sympathize with your criticism of projects that amount to "if you randomly bash it, it breaks", particularly in vertebrate neuroscience where these seem to get published most often. But let me say why I don't think this paper is simply another of those.
First, the idea that a single gene change — or even a single allele swap — could cause drastic behavioral changes is not always as far out as you (and Mark Liberman) imply, although it is certainly the exception not the rule. Two extreme examples come to mind — monogamy in voles and courtship behavior in fruit flies. (There are quite a few other examples.) It's true that it is unscientific to automatically claim there's a "gene for X"; but so is discarding the idea out of hand.
[(myl) Alas, the vole monogamy stuff is almost as over-sold as the "language gene" stuff — see here. I don't know about the fruitfly courtship work. ]
Second, if you look at the paper, I think you will find your criticism — that they were looking for mice to acquire syntax when they should have been looking at how Foxp2 affects neuronal development — to be out of line. Of the paper's six figures, one is a technique figure, two are behavioral (exploratory behavior and vocal calls), and three are about effects of Foxp2 on neuronal development and function (dopamine production, dendrite length, LTD). There is a long-ish line of evidence pointing to an important role for Foxp2 in language; this paper mostly looks at what Foxp2 does for neurons.
[(myl) Yes, I agree. But as I noted in commenting on the earlier comment, this particular experiment would probably never have been done without the "language gene" buzz. And I expect that the experimenters hoped for a more striking effect on the vocalizations — though why they'd expect it to show up in newborns, rather than as an enhancement of vocal learning possibilities, is another interesting question. ]
Nathan Myers said,
June 4, 2009 @ 5:51 pm
I'm curious about this FOXP2 vs Foxp2 notation. It seems akin to gender, or capitalizing the monotheistic "Him", as a way to concisely forestall an otherwise easy confusion. Is it common to discuss human and (other-) animal genes in the same paper, and this makes it easier to keep straight which is which? Or have largely separate literatures adopted different conventions that are colliding in this instance?
When such a paper is read, is FOXP2 pronounced any differently from Foxp2? I'm thinking of Brian Kernighan's "telephone test" ( http://www.eecs.harvard.edu/~ellard/CS50-95/programming-style.html , #2) by which program variable names should be pronounceable, and distinguishable from one another when read over the telephone.
mgh said,
June 4, 2009 @ 6:55 pm
Mark, From your own post on the vole research: "My conclusions: The vole pair-bonding research is terrific stuff, which leaves a lot of fascinating open questions." and then you go on to criticize the overinterpretation fo the human work.
[(myl) The vole work, as I understand it, shows that arginine vasopressin levels in the lateral septum of rodents play an essential role in social recognition, and that under- or over-expression of the V1aR gene in that region of the brain can push those levels up and down. Animals whose lateral septum is deficient in this hormone can't recognize conspecific individuals, which of course inhibits pair bonding. But this is really about social recognition more than pair bonding. And it's not about a mutation in a particular gene, as far as I know, but rather about the level of expression of a particular gene (or the resulting levels of the AVP hormone) in a particular part of the brain. Presumably some more extensive genetic circuit is involved in the modulation of that gene's expression in that specific brain structure in males of certain rodent species.
If I've got this right, then it's over-interpretation to refer to V1aR as "the pair-bonding gene" — and roughly in the same way as referring to FOXP2 as "the language gene". And I wouldn't be surprised to learn that the situation is more complicated than I know, rather than simpler. ]
The fruit fly work is very solid — essentially a splice variant toggles between the male and female versions of large chunks of the courtship program — but does not relate to a variation found in nature.
I can post a short list of other examples if you're interested (mouse sex discrimination for starters). Although, by all means, finding a gene that even weakly explains big chunks of behavioral programs is still a major accomplishment, and usually it is not that simple.
I think it's important to separate criticism of the buzz from criticism of the scientists' own interests. Human p53, huntingtin, and many others have been knocked in to mice, so it's not that unusual an approach, and most mouse geneticists are pretty savvy about the limits of interpreting mouse models.
Nathan Myers, I see why you might think that but the nomenclature for each field arose on its own and I wouldn't read too much into it. For budding yeast, genes are all caps italics (YFG1) unless it's a recessive mutation (yfg1) and proteins are initial cap roman (Yfg1). For nematodes, genes are lower case italics with a hyphen (yfg-1) and proteins are all caps roman (YFG-1). It gets confusing when you are talking about the same gene across species and need to keep all the nomenclature straight, but except for reviews it doesn't come up that much. They are all pronounced the same way (in this case, "Fox pee two").
Language Log » Mice with the “language gene” stay mum « blagorama said,
June 4, 2009 @ 7:37 pm
[…] See the original post: Language Log » Mice with the “language gene” stay mum […]
Jesse Hochstadt said,
June 4, 2009 @ 9:02 pm
Your claim that 'mice with the human "language gene" didn't chatter — they just squeaked a little more gruffly and plainly than their wild-type littermates' is incorrect. You've omitted the following finding: "calls with frequency jumps lasted longer (ANOVA, n = 149, p < 0.05), had longer gaps (p < 0.05) and started (p < 0.01) and ended (p < 0.05) with higher peak frequencies in Foxp2hum/hum mice than in their wild-type littermates." So while the results hardly show that the mice "chatter," neither do they show that genetic change absolutely lowers pitch or decreases pitch modulation, as might be the case if it, say, simply (as Tim Silverman suggests) "damaged" the ability to vocalize.
[(myl) Sorry for missing that — I've edited the body of the text to correct the error. I'm not sure whether to interpret those differences as "damage" or not, though, because you could take the fact that the calls with jumps lasted longer and had longer gaps to mean that they were just performed more slowly.
The authors say that "the full [vocalization] dataset is available on request", and in fact I'd like to take a look at it. It's worth pointing out that none of the effects were especially large ones — thus in the case of the calls with frequency jumps, the h/h mice had a mean peak frequency start of 66.9 hKhz, compared to 63.9 kHz for the wt/wt mice, a difference of around 5% (though I estimate from the S.E. measures that this is roughly 0.4 of a standard deviation, since each strain's calls are apparently pretty stereotyped). ]
Also, Enard et al. report numerous results showing that mice heterozygous for the wild-type gene and a knockout version of that gene, which "show intermediate levels of Foxp2 protein … and can thus be used to assess the consequences of reduced Foxp2 expression," show effects different from and often in the opposite direction, relative to homozygous wild-type mice, from those in the "humanized" mice.
None of this shows, of course, that the "humanized" mice have a "parser" or, more broadly, the capacity for complex sequencing that characterizes human motoric and cognitive ability. Nor do Enard et al. discuss the ethological significance (if any) of the different call types they analyze, or any behavioral changes of the mothers in response to the altered isolation cries (Enard et al. analyzed "ultrasonic vocalizations emitted by pups when placed outside the nest"). But the vocalization results aren't quite as simple as you indicate. (It's also worth noting that Enard et al. provide a properly cautious discussion of the limitations of their results with respect to making inferences about the evolution of language.)
[(myl) The discussion in the paper is a model for careful, scrupulous presentation of an experiment that could easily have been over-interpreted. I think that's probably why the paper has not gotten more media play, unfortunately. ]
(Full disclosure: Philip Lieberman was my doctoral adviser and I did research with him on the effects of Parkinson's disease, a primarily basal ganglia disorder, on the ability to comprehend syntactically complex sentences as well as on other aspects of complex cognition.)
Chris said,
June 5, 2009 @ 9:29 am
Surely one of the best examples of a "gene for X" is SRY, the gene for maleness in therians. Embryos that have it will develop as male and embryos that lack it will develop as female, both with very high reliability (although there are rare exceptions).
Of course, the details of male or female development depend on lots of other genes, but almost all determination of *which* development path is followed depends on the presence or absence of SRY.
Most other genes aren't quite that clear-cut in their effects, though – both the male and female developmental pathways of any given therian species have coexisted and coevolved for tens of millions of years.
[(myl) Of course there are cases where specific aspects of the genotype line up with specific aspects of the phenotype. The problem with the "gene for X" idea is that many people — including, apparently, most headline writers, many science journalists, and some scientists — think that if there is a salient phenotypic feature or dimension, there must a gene that corresponds. ]
Mark F. said,
June 5, 2009 @ 11:03 am
I think you're pretty hard on Wade. Even the headline says "A Language Gene", not "The Language Gene", which looks to me like it might not be so far off. The article gives enough background to explain why the gene is associated to language, so that readers can tell for themselves that it can't be "the gene for language", since people with broken versions of the gene still have some language abilities. And he does say it's "perhaps surprising" that giving mice a human-like version of the gene changes their vocalizations even at all. If that's true, then of course it'd be "completely and totally unexpected" if they got a talking mouse.
[(myl) As I hope the context makes clear, I based my evaluation of Nicholas Wade's role in promoting the "language gene" idea on the half-dozen or so other stories of his that I linked to, including this one, where he wrote:
]
BMH said,
June 7, 2009 @ 10:33 am
Are the conclusions about how this has affected the 'speech' abilities of mice premature? That is, given that FoxP2 is involved in language in humans, it presumably is involved in giving us language acquiring abilities and not language itself. Human pups still have to mature for a few years and be exposed to the right stimuli to get to recursive linguistic ability, right? Why not the same for these mice pups? Isn't there a broad range of developmental experiments to be done here, exposing the mice to a variety of linguistically-related stimuli to see what cognitive capacities this genetic change has had on them?
I'm kind of disappointed that this hasn't been noted.