When more data makes things worse…
« previous post | next post »
The mantra of machine learning, as Fred Jelinek used to say, is "The best data is more data" — because in many areas, there's a Long Tail of relevant cases that are hard to classify or predict without either a valid theory or enough examples.
But a recent meta-analysis of machine-learning work in digital medicine shows, convincingly, that more data can lead to poorer reported performance. The paper is Visar Berisha et al., "Digital medicine and the curse of dimensionality", NPJ digital medicine 2021, and one of the pieces of evidence they present is shown in the figure reproduced below:
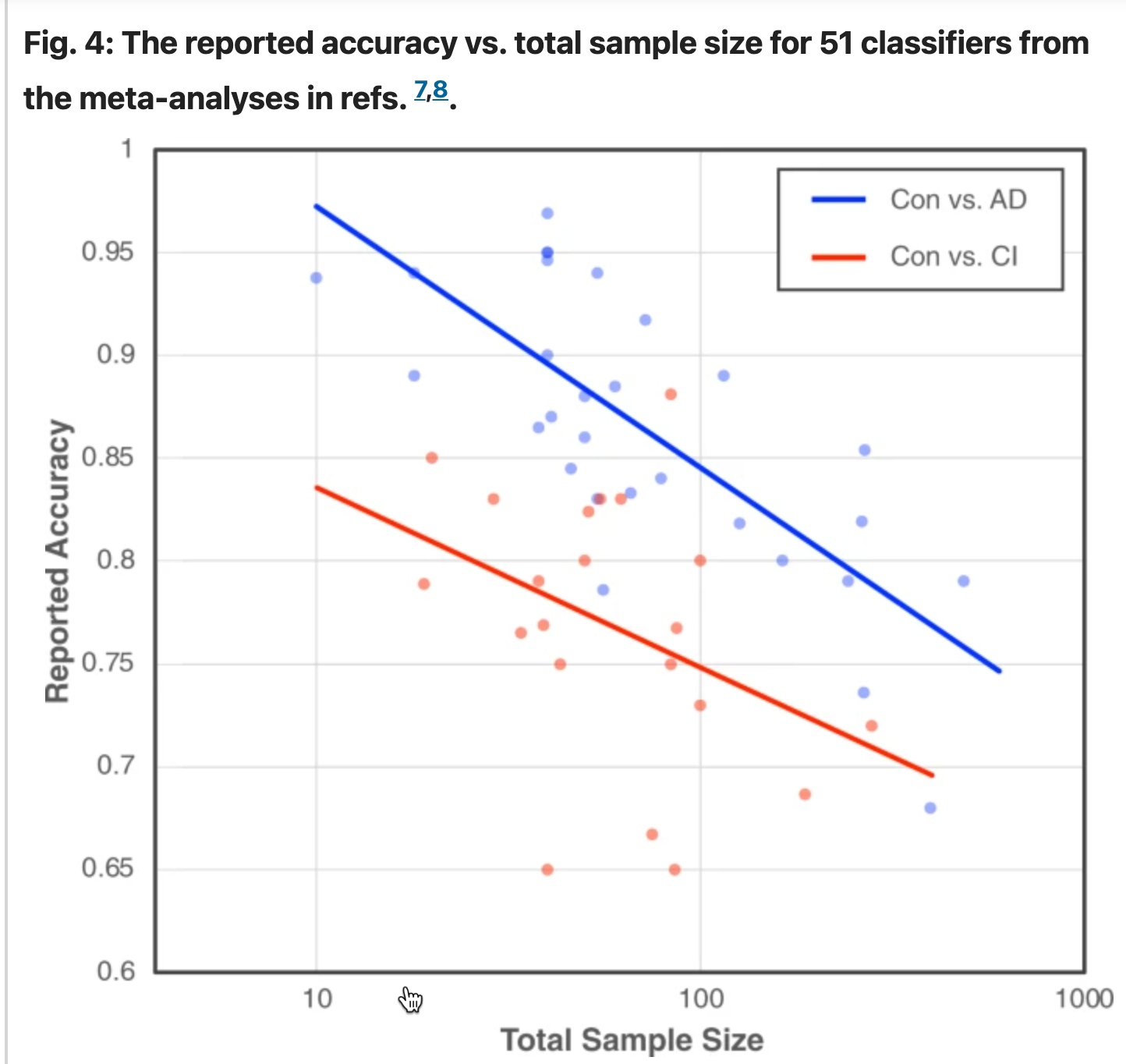
This analysis considers two types of models: (1) speech-based models for classifying between a control group and patients with a diagnosis of Alzheimer’s disease (Con vs. AD; blue plot) and (2) speech-based models for classifying between a control group and patients with other forms of cognitive impairment (Con vs. CI; red plot).
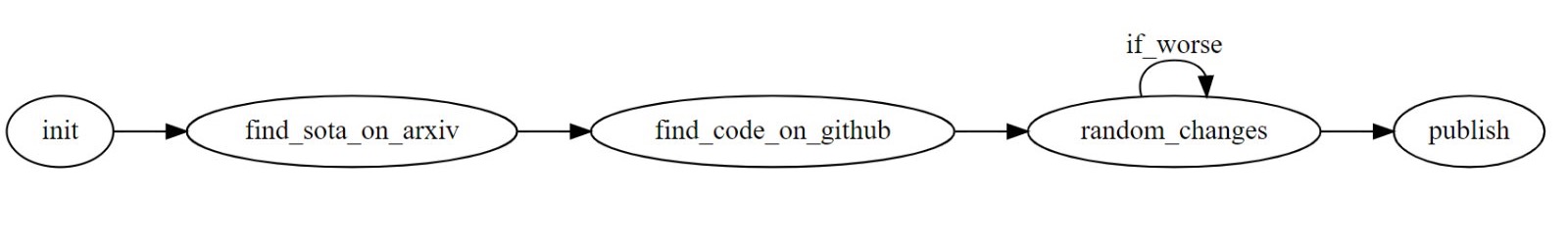
This effect is basically a form of "Graduate Student Descent", a semi-pun on the optimization method of "gradient descent". The description in this 2020 article offers a flow chart for this method:
The "random_changes" are not, of course, always random. Changes in the machine-learning architecture, or additional "features" derived from the available data, may well be based on plausible hypotheses. But such explorations necessarily lead to some amount of over-fitting. And smaller datasets lead to more over-fitting, and therefore to better estimated performance and worse generalization to new observations.
A maximally simple simulation illustrates this effect. We create 50 random features for each of a set of N "subjects", and look at the performance of plain old linear discriminant analysis, as N varies from 50 to 800:
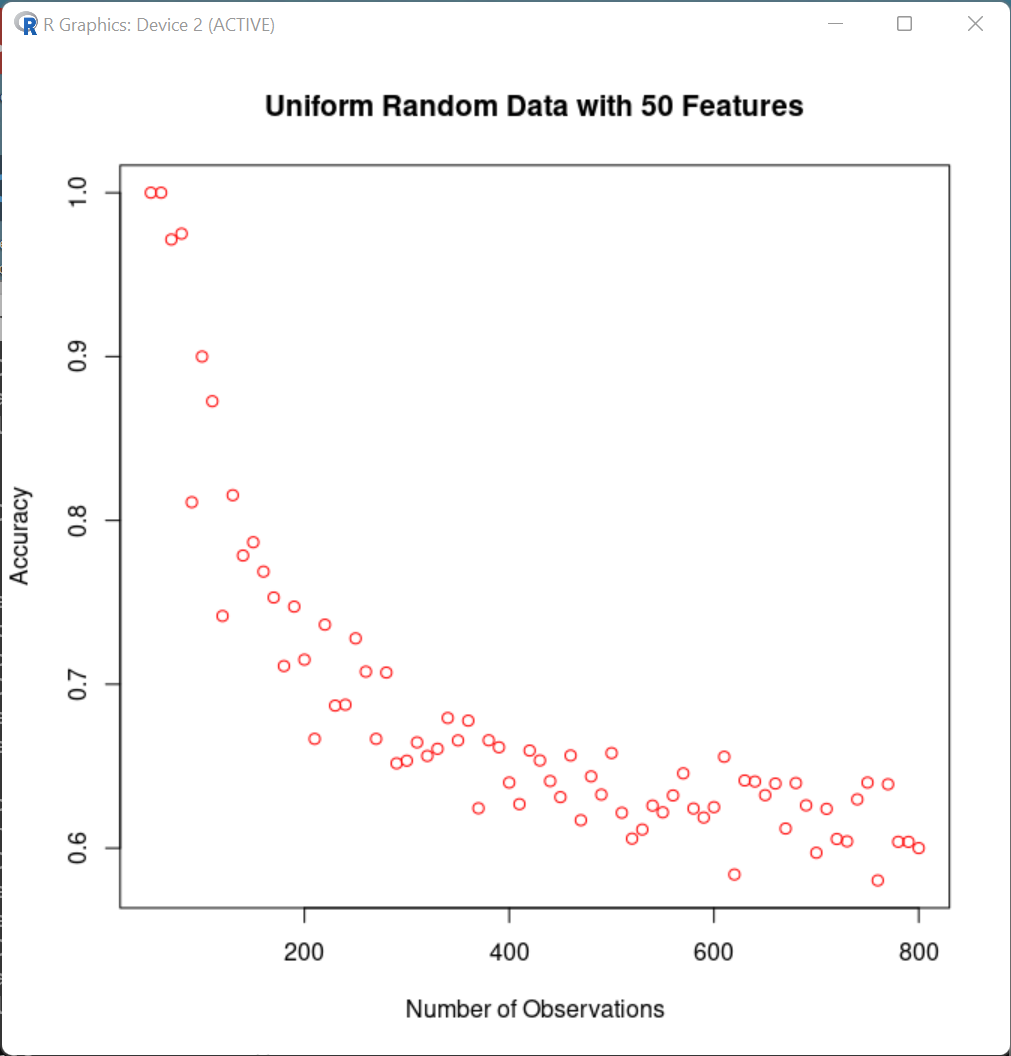
(R code is here.)
More realistic simulations would involve leave-one-out cross validation (or some other train/test division), various more sophisticated ML methods, etc. But the same basic pattern emerges — it's easier to fool yourself (and others) when your system is classifying a smaller number of observations.
Adding more random "features" for a given number of subjects generally yields a similar effect.
How can we avoid this? In the foundational work on Human Language Technology, roughly from 1985 to 2010, the practice was to keep "Evaluation Test" data secret from researchers until final testing time; use it only once (at least ideally); and change the dataset and the task details regularly. And even more important, the dataset sizes were large enough that the simplest kinds of over-fitting were avoided.
A key problem with clinical applications is that the dataset sizes are typically tiny, due to the culture of the field, which uses genuine concerns about privacy and confidentiality to protect more selfish motives in protecting each group's proprietary data. This makes it very hard to avoid the kind of over-fitting illustrated above. And another consequence is that each dataset tends to have its own set of locally-specific uncontrolled covariates.
So in conclusion, it's still true that more data is better — and the fact that getting data from more subjects can lead to lower estimated performance is actually evidence for the value of getting data from more subjects.
unekdoud said,
August 29, 2022 @ 9:19 am
Applying this observation to itself: you might overestimate your model's ability to generalize if you don't test for it.
mg said,
August 29, 2022 @ 9:34 am
The problem of over-fitting is one that statisticians are very aware of – unfortunately, too much of this research is done without input of statisticians (people who've just had a couple of courses on machine learning don't count). We're very aware of issues of the need for large numbers of participants not just large numbers of utterances – 100 total utterances from 4 people does not give as much data as 100 utterances from 25, since there's within-person correlation.
Yuval said,
August 29, 2022 @ 10:33 am
Not sure I follow the claims regarding HLT/NLP data. Once a shared task like CoNLL (2003, 2006, 2010 etc.) was finished, test sets were released and any future research was done with them available. The WSJ PTB with its "standard split" was AFAIK always fully available since 1993. If anything, it's benchmarks like GLUE and SuperGLUE from the late 2010's that started hiding test set labels, or entire test sets, theoretically in perpetuity, only offering an API of "gimme system, get score, we publish your score whether you like it or not".
[(myl) My reference was to DARPA shared tasks, which began in the late 1980s. CoNLL was a sort of bottom-up civilian copy, which was not managed in the same way. It's false that GLUE and SuperGLUE were the first to hide evaluation test sets. DARPA and NIST always did that, and also changed them for every evaluation, even if there were retrospective evaluations for comparison purposes as well.]
Kris said,
August 29, 2022 @ 3:30 pm
Lots to unpack here. First, as you say, more data is always better, lower reported accuracy notwithstanding. Lower in this instance basically meaning a more realistic estimate of actual accuracy.
I realize accuracy is probably about as good as the researcher had to work with given it was a meta analysis of published papers and needed a metric that lots of papers report. But accuracy is generally a poor way of evaluating classifiers to begin with. Unless all the classifiers were trained on balanced samples – which is really about the only situation I'd consider accuracy a halfway decent evaluation metric – or at least similarly unbalanced samples (impossible to imagine), then this tells us very little in and of itself. Maybe the small sample sizes tended to have unbalanced training samples and all the large sample sizes tended to have balanced training samples.
[(myl) Indeed — in a dataset where there are twice as many cases of group A as group B, the baseline performance should be 67%, not 50%.]
It's easy to construct a scenario where classifier A has a higher accuracy than classifier B, even though the true positive rate and the true negative rate are both better for classifier B! AND even when both classifiers are built on balanced samples! A nice corollary illustration of Simpson's paradox.
More to the point, a classifier could have perfect rank ordering of individuals, but a lackluster accuracy, because accuracy takes a hard arbitrary cutoff and says above this line is a 1 and below is a 0. Two people with Alzheimer's, one just beginning to exhibit measurable symptoms and one 20 years advanced into the disease, and a clinician has diagnosed both as having Alzheimer's. Our classifier gives the first a score of 0.48, the second a score of 0.95, when the "real" values of their clinical degree of disease progression are 0.5 and 0.95 (assume). Our classifier is incredibly good, but accuracy is terrible because 0.5 is the arbitrary cutoff.
[(myl) Again agreed, which is the motivation behind AUC and similar evaluation methods.]
Moral – you probably don't meaningfully evaluate a model by accuracy (except maybe in constrained and unrealistic circumstances), and if you do, be prepared for unintuitive results on model comparisons. Second, you don't do a study on 10 data points (how did that get published anywhere?), and if you do, model evaluation means nothing. A simulation that shows what happens when you predict 50 samples using 50 covariates is a whole other matter which is related, but any study that does anything close to that should be rejected by reviewers out of hand.
Last, it's a lot easier to get high accuracy when your sample is small because it is highly selective. First, let me throw out all the samples where Alzheimer's is a new diagnosis. Then throw out all the females, then all the people not between ages 55-70, then let's just keep the majority race to eliminate that confounder, throw out low income and non-adherent individuals, and I've got a nice clean sample of a few dozen remaining. Nice good accuracy on this model. Until I try to apply my model to any new sample that it didn't train on. It will not generalize to any random sample I could ever pull, and is thus useless for everyone who is not a high income adherent male between 55-70 with either no diagnosis or a long-standing diagnosis of Alzheimer's. In other words, just plain useless. Especially if I'm trying to compare the "performance" of this model (judged by accuracy, again) to that of other models.
[(myl) Which leads into the whole precision/recall/F1/PPV etc. business… See e.g. "Steven D. Levitt: pwned by the base rate fallacy?", 4/10/2008; When 90% is 32%", 3/18/2014; or these lecture notes on "Scoring Binary Classification" from a 2014 course.]
Michael Watts said,
August 29, 2022 @ 11:31 pm
I don't think this is right. You can achieve 67% fairly easily using a strategy of "everything is A". But how do you choose that strategy? First you'd have to successfully learn that A is more common than B; without that, you're equally likely to decide that "everything is B" and achieve 33% accuracy. Baseline accuracy is still 50%.
[(myl) So would you take my bet, at even odds, that your lottery ticket won't win? Please do!
We're talking about Bayesian methods of machine learning, in situations where testing material is generally similar to training material. And we know what the overall rates are in the training material.]
Michael Watts said,
August 30, 2022 @ 10:59 am
Are we? This is what I see us talking about:
Kris said,
August 30, 2022 @ 3:04 pm
The simplest possible classifier is one that predicts all observations to be the majority class (called a naive classifier sometimes). The accuracy of this model is, by definition, whatever the proportion of the majority class is in the population. If you are training on a representative sample of your population, then your model needs to be beat this naive classifier on accuracy or your model is trash. Mark is exactly correct as he describes it above. If there are two As for every B in the population, your model accuracy needs to be better than 67% to be worth anything, because the dumbest reasonable classifier imaginable can achieve 67% accuracy. If X and Y are equally likely in the population, your model has a more difficult task (ceteris paribus) and needs to beat a baseline accuracy of 50% before you would consider it.
So, if you compare a model trying to discriminate A from B to a model trying to discriminate X from Y, simple accuracy will not tell you which model is doing a better job. Which is why training sample balance was a point of discussion in my original comment – each of these classifiers might be using a different sample balance of classes for training, which muddies the comparisons of performance beyond the other pitfalls of using accuracy which exist even if they were all identically balanced.
James Wimberley said,
August 31, 2022 @ 7:43 am
I wish I could follow this debate, but my life expectancy is now too low to follow Solon's advice to "keep learning new things" in this instance.
As an aside: "A key problem with clinical applications is that the dataset sizes are typically tiny, due to the culture of the field, which uses genuine concerns about privacy and confidentiality to protect more selfish motives in protecting each group's proprietary data."
Medical science does now include examples of research done by crunching vast samples from increasingly integrated databases, for instance this, with n>12 million. I Am Not A Statistician but suppose that the problem with such huge samples is not getting high statistical significance in a correlation but more the temptation to chase trivial effect sizes down rabbit-holes.